# install.packages("pak")
# pak::pak("tidymodels", "embed", "vetiver", "pins", "smdocker")
library(tidymodels)
library(embed)
library(vetiver)
library(pins)
library(smdocker)AWS, CSV Input, SageMaker Endpoint
This page was last generated on 2024-02-29. If you find the code out of date please file an issue.
All changes from the standard pipeline are highlighted with a cranberry line to the right.
Loading packages
We are using the tidymodels package to do the modeling, embed for target encoding, pins for versioning, vetiver for version and deployment, and smdocker for deploying to SageMaker.
Loading Data
We are using the smaller laxflights2022 data set described on the data preparation page.
flights <- readr::read_csv(here::here("data/laxflights2022_lite.csv"))
glimpse(flights)Rows: 3,757
Columns: 8
$ arr_delay <dbl> 4, -15, -12, 38, -9, -17, 5, 12, -40, 6, -7, 28, 25, -9, 180…
$ dep_delay <dbl> 9, -8, 0, -7, 3, 6, 29, -1, 2, 7, 6, 13, 34, -2, 191, 52, 9,…
$ carrier <chr> "UA", "OO", "AA", "UA", "OO", "OO", "UA", "AA", "DL", "DL", …
$ tailnum <chr> "N37502", "N198SY", "N410AN", "N77261", "N402SY", "N509SY", …
$ origin <chr> "LAX", "LAX", "LAX", "LAX", "LAX", "LAX", "LAX", "LAX", "LAX…
$ dest <chr> "KOA", "EUG", "HNL", "DEN", "FAT", "SFO", "MCO", "MIA", "OGG…
$ distance <dbl> 2504, 748, 2556, 862, 209, 337, 2218, 2342, 2486, 862, 156, …
$ time <dttm> 2022-01-01 13:15:00, 2022-01-01 14:00:00, 2022-01-01 14:45:…Modeling
As a reminder, the modeling task we are trying to accomplish is the following:
Given all the information we have, from the moment the plane leaves for departure. Can we predict the arrival delay
arr_delay?
Our outcome is arr_delay and the remaining variables are predictors. We will be fitting an xgboost model as a regression model.
Splitting Data
Since the data set is already in chronological order, we can create a time split of the data using initial_time_split(), this will put the first 75% of the data into the training data set, and the remaining 25% into the testing data set.
set.seed(1234)
flights_split <- initial_time_split(flights, prop = 3/4)
flights_training <- training(flights_split)Since we are doing hyperparameter tuning, we will also be creating a cross-validation split
flights_folds <- vfold_cv(flights_training)Feature Engineering
We need to do a couple of things to make this data set work for our model. The datetime variable time needs to be transformed, as does the categorical variables carrier, tailnum, origin and dest.
From the time variable, the month and day of the week are extracted as categorical variables, then the day of year and time of day are extracted as numerics. The origin and dest variables will be turned into dummy variables, and carrier, tailnum, time_month, and time_dow will be converted to numerics with likelihood encoding.
flights_rec <- recipe(arr_delay ~ ., data = flights_training) %>%
step_novel(all_nominal_predictors()) %>%
step_other(origin, dest, threshold = 0.025) %>%
step_dummy(origin, dest) %>%
step_date(time,
features = c("month", "dow", "doy"),
label = TRUE,
keep_original_cols = TRUE) %>%
step_time(time, features = "decimal_day", keep_original_cols = FALSE) %>%
step_lencode_mixed(all_nominal_predictors(), outcome = vars(arr_delay)) %>%
step_zv(all_predictors())Specifying Models
We will be fitting a boosted tree model in the form of a xgboost model.
xgb_spec <-
boost_tree(
trees = tune(),
min_n = tune(),
mtry = tune(),
learn_rate = 0.01
) %>%
set_engine("xgboost") %>%
set_mode("regression")xgb_wf <- workflow(flights_rec, xgb_spec)Hyperparameter Tuning
doParallel::registerDoParallel()
xgb_rs <- tune_grid(
xgb_wf,
resamples = flights_folds,
grid = 10
)i Creating pre-processing data to finalize unknown parameter: mtryWe can visualize the performance of the different hyperparameter selections
autoplot(xgb_rs)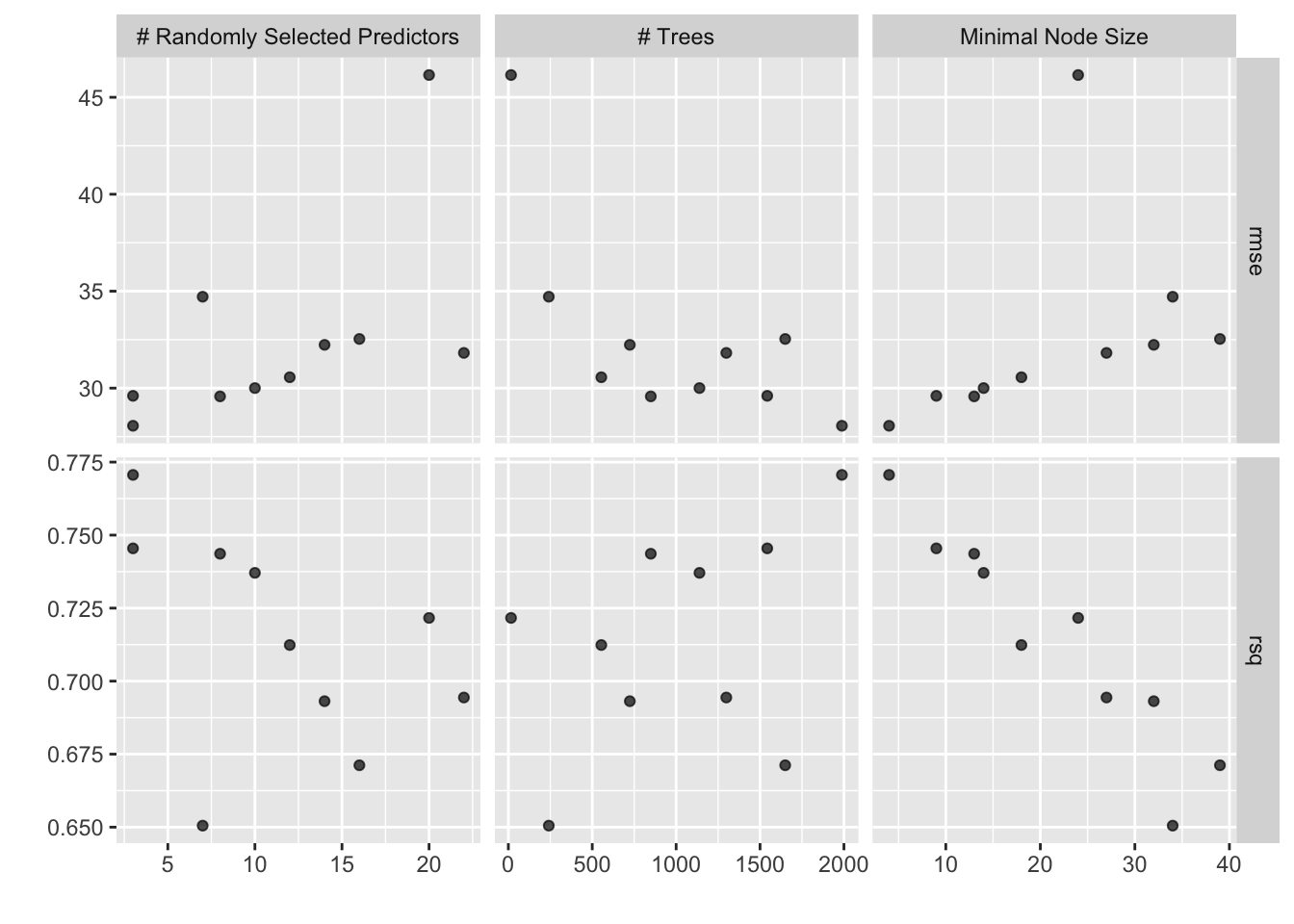
and look at the top result
show_best(xgb_rs, metric = "rmse")# A tibble: 5 × 9
mtry trees min_n .metric .estimator mean n std_err .config
<int> <int> <int> <chr> <chr> <dbl> <int> <dbl> <chr>
1 3 1988 4 rmse standard 28.1 10 5.88 Preprocessor1_Model01
2 8 849 13 rmse standard 29.6 10 6.44 Preprocessor1_Model04
3 3 1543 9 rmse standard 29.6 10 6.11 Preprocessor1_Model02
4 10 1139 14 rmse standard 30.0 10 6.44 Preprocessor1_Model05
5 12 554 18 rmse standard 30.6 10 6.77 Preprocessor1_Model06Fitting Final Model
Once we are satisfied with the modeling that has been done, we can fit our final model. We use finalize_workflow() to use the best hyperparameters, and last_fit() to fit the model to the training data set and evaluate it on the testing data set.
xgb_last <- xgb_wf %>%
finalize_workflow(select_best(xgb_rs, metric = "rmse")) %>%
last_fit(flights_split)Creating vetiver model
v <- xgb_last %>%
extract_workflow() %>%
vetiver_model("flights_xgb")
v
── flights_xgb ─ <bundled_workflow> model for deployment
A xgboost regression modeling workflow using 7 featuresVersion model with pins on Amazon SageMaker
We will version this model on Amazon S3 using the pins package.
For the smoothest experience, we recommend that you authenticate using environment variables. The two variables you will need are AWS_ACCESS_KEY_ID and AWS_SECRET_ACCESS_KEY.
Depending on your S3 setup, you will need to use additional variables to connect. Please see https://github.com/paws-r/paws/blob/main/docs/credentials.md and this pins issue for help if the following paragraphs doesn’t work for you.
The function usethis::edit_r_environ() can be very handy to open .Renviron file to specify your environment variables.
You can find both of these keys in the same location.
- Open the AWS Console
- Click on your username near the top right and select
Security Credentials - Click on
Usersin the sidebar - Click on your username
- Click on the
Security Credentialstab - Click
Create Access Key - Click
Show User Security Credentials
Once you have those two, you can add them to your .Renviron file in the following format:
AWS_SECRET_ACCESS_KEY=xxxxxxxxxxxxxxxxxxxxxxxxxxxx
AWS_ACCESS_KEY_ID=xxxxxxxxxxxxxxxxxxxxxxxxxxxxNote that you don’t want to put quotes around the values.
Once that is all done, we can create a board that connects to Amazon S3, and write our vetiver model to the board. Now that you have set up the environment variables, we can create a pins board. When using S3 you need to specify a bucket and its region. This cannot be done with Pins and has to be done beforehand.
board <- board_s3(
"tidymodels-pipeline-example",
region = "us-west-1"
)
vetiver_pin_write(board, v)Since we are using vetiver_deploy_sagemaker() which uses the {smdocker} package, we need to make sure that we have the right authetication and settings.
If you are working locally, you will likely need to explicitly set up your execution role to work correctly. Check out Execution role requirements in the smdocker documentation, and especially note that the bucket containing your vetiver model needs to be added as a resource in your IAM role policy.
Once we are properly set up, we can use vetiver_deploy_sagemaker(), it takes a board, the name of endpoint and the instance_type Look at the Amazon SageMaker pricing to help you decide what you need. Depending on your model, it will take a little while to run as it installs what it needs.
new_endpoint <- vetiver_deploy_sagemaker(
board = board,
name = "flights_xgb",
instance_type = "ml.t2.medium"
)Make predictions from Connect endpoint
With the endpoint we can pass in some data set to predict with.
predict(
new_endpoint,
flights_training
)# A tibble: 2,817 × 1
.pred
<dbl>
1 -1.79
2 -13.3
3 -17.3
4 -3.67
5 82.8
6 52.2
7 10.7
8 8.04
9 58.1
10 5.39
# ℹ 2,807 more rows:::