tidyclust
expanding tidymodels to clustering
tidymodels
- Consistent
- Modular
- Extensible

![]()
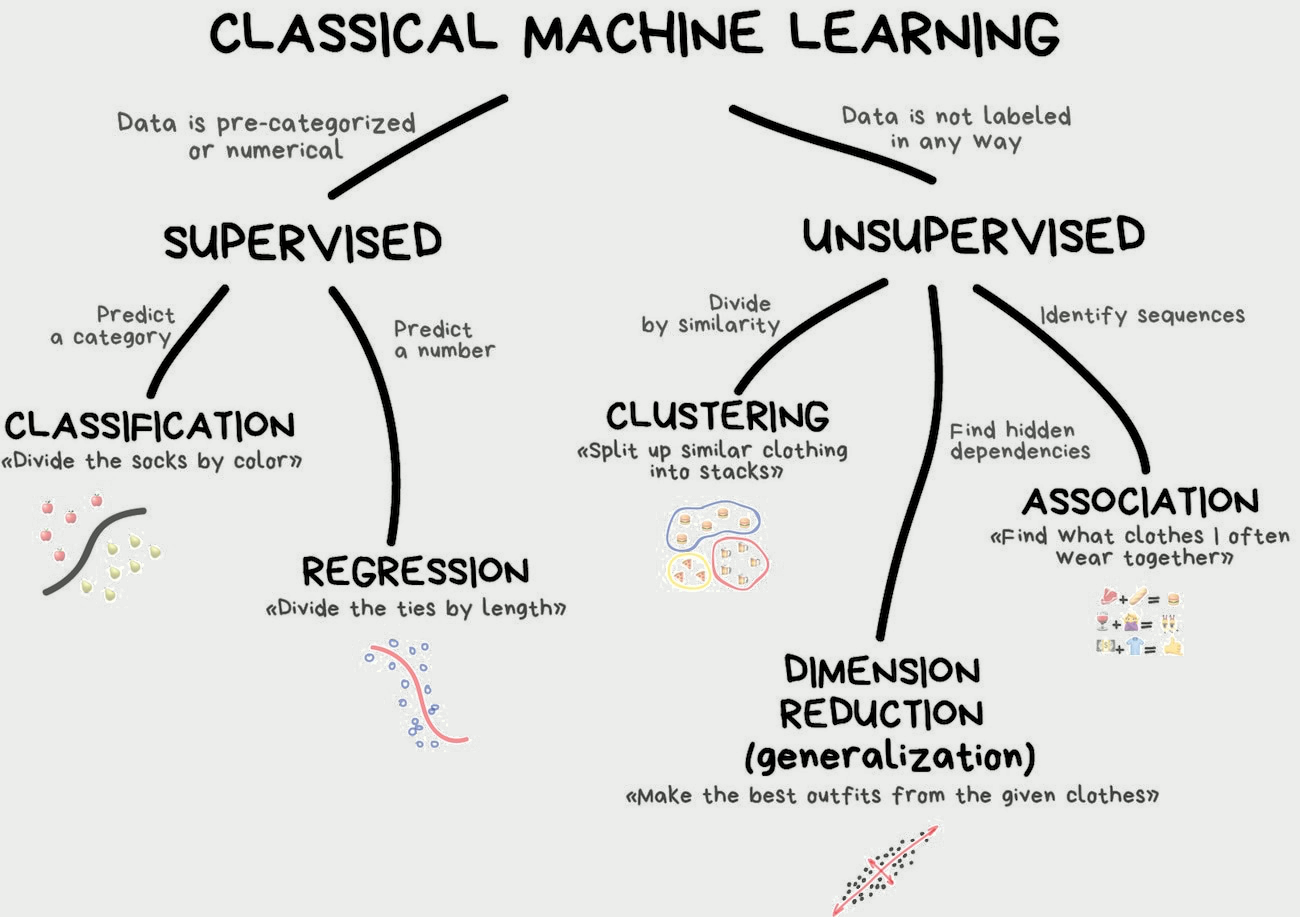
Illustration credit: https://vas3k.com/blog/machine_learning/
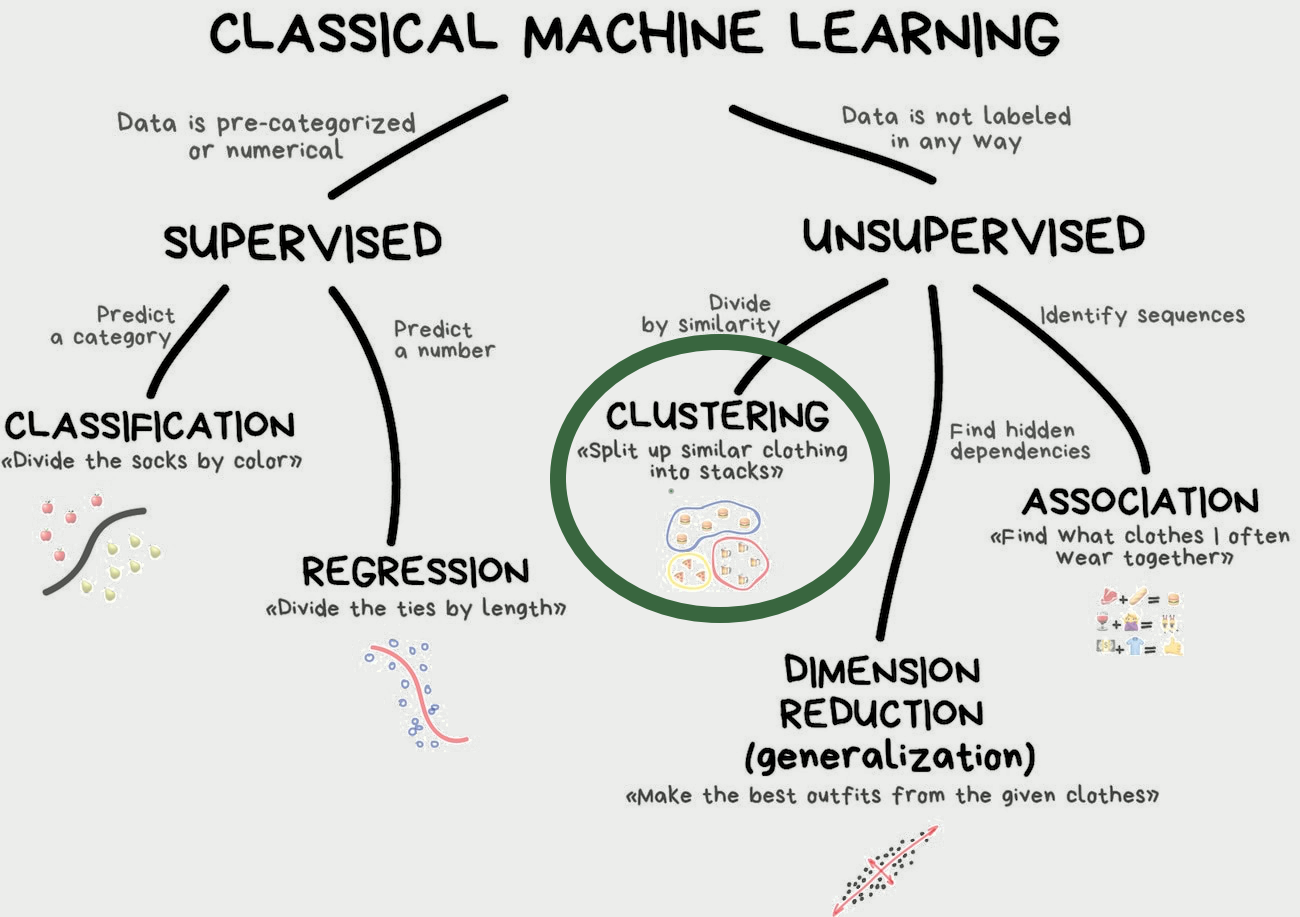
Illustration credit: https://vas3k.com/blog/machine_learning/
T
I
D
Y
C
L
U
S
T
![]()
![]()
![]()
T
I
D
Y
C
L
U
S
T
![]()
![]()
![]()
Why another package?
![]()
tidymodels was build with supervised models in mind
tidymodels
Models have outcomes
Clearly defined predict()
Easy to estimate performance
tidyclust
Models don’t have outcomes
No clearly defined predict()
No clear answer
![]()
![]()
Why another package?
![]()
![]()
There is some reimplementation in tidyclust
Experience will be as seamless as possible
Date Fruit Data
glimpse(dates)
#> Rows: 898
#> Columns: 34
#> $ area <dbl> 422163, 338136, 526843, 41…
#> $ perimeter <dbl> 2378.908, 2085.144, 2647.3…
#> $ major_axis <dbl> 837.8484, 723.8198, 940.73…
#> $ minor_axis <dbl> 645.6693, 595.2073, 715.36…
#> $ eccentricity <dbl> 0.6373, 0.5690, 0.6494, 0.…
#> $ eqdiasq <dbl> 733.1539, 656.1464, 819.02…
#> $ solidity <dbl> 0.9947, 0.9974, 0.9962, 0.…
#> $ convex_area <dbl> 424428, 339014, 528876, 41…
#> $ extent <dbl> 0.7831, 0.7795, 0.7657, 0.…
#> $ aspect_ratio <dbl> 1.2976, 1.2161, 1.3150, 1.…
#> $ roundness <dbl> 0.9374, 0.9773, 0.9446, 0.…
#> $ compactness <dbl> 0.8750, 0.9065, 0.8706, 0.…
#> $ shapefactor_1 <dbl> 0.0020, 0.0021, 0.0018, 0.…
#> $ shapefactor_2 <dbl> 0.0015, 0.0018, 0.0014, 0.…
#> $ shapefactor_3 <dbl> 0.7657, 0.8218, 0.7580, 0.…
#> $ shapefactor_4 <dbl> 0.9936, 0.9993, 0.9968, 0.…
#> $ mean_rr <dbl> 117.4466, 100.0578, 130.95…
#> $ mean_rg <dbl> 109.9085, 105.6314, 118.57…
#> $ mean_rb <dbl> 95.6774, 95.6610, 103.8750…
#> $ std_dev_rr <dbl> 26.5152, 27.2656, 29.7036,…
#> $ std_dev_rg <dbl> 23.0687, 23.4952, 24.6216,…
#> $ std_dev_rb <dbl> 30.1230, 28.1229, 33.9053,…
#> $ skew_rr <dbl> -0.5661, -0.2328, -0.7152,…
#> $ skew_rg <dbl> -0.0114, 0.1349, -0.1059, …
#> $ skew_rb <dbl> 0.6019, 0.4134, 0.9183, 1.…
#> $ kurtosis_rr <dbl> 3.2370, 2.6228, 3.7516, 5.…
#> $ kurtosis_rg <dbl> 2.9574, 2.6350, 3.8611, 8.…
#> $ kurtosis_rb <dbl> 4.2287, 3.1704, 4.7192, 8.…
#> $ entropy_rr <dbl> -59191263232, -34233065472…
#> $ entropy_rg <dbl> -50714214400, -37462601728…
#> $ entropy_rb <dbl> -39922372608, -31477794816…
#> $ al_ldaub4rr <dbl> 58.7255, 50.0259, 65.4772,…
#> $ al_ldaub4rg <dbl> 54.9554, 52.8168, 59.2860,…
#> $ al_ldaub4rb <dbl> 47.8400, 47.8315, 51.9378,…Date Fruit Data
![]()
Specifying a clustering model
![]()
Specifying a clustering model
![]()
Specifying a clustering model
![]()
cluster assignment + clusters + prediction
![]()
extract_cluster_assignment(kmeans_fit)
#> # A tibble: 898 × 1
#> .cluster
#> <fct>
#> 1 Cluster_1
#> 2 Cluster_2
#> 3 Cluster_1
#> 4 Cluster_2
#> 5 Cluster_2
#> 6 Cluster_2
#> 7 Cluster_1
#> 8 Cluster_2
#> 9 Cluster_1
#> 10 Cluster_1
#> # … with 888 more rows
#> # ℹ Use `print(n = ...)` to see more rowscluster assignment + clusters + prediction
![]()
extract_centroids(kmeans_fit)
#> # A tibble: 5 × 7
#> .cluster PC1 PC2 PC3 PC4 PC5
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 Cluster_1 2.15 3.18 1.42 0.0640 -0.342
#> 2 Cluster_2 -5.66 -1.50 0.806 0.298 -0.361
#> 3 Cluster_3 3.80 -2.82 -0.786 -0.159 -0.00248
#> 4 Cluster_4 0.934 -0.774 -0.102 0.498 0.0410
#> 5 Cluster_5 -1.99 3.37 -2.05 -1.02 1.06
#> # … with 1 more variable: PC6 <dbl>
#> # ℹ Use `colnames()` to see all variable namescluster assignment + clusters + prediction
![]()
Metrics
![]()
Metrics
![]()
tuning
![]()
tuning
![]()
tuning
![]()
collect_metrics(res)
#> # A tibble: 10 × 7
#> num_clus…¹ .metric .esti…² mean n std_err
#> <int> <chr> <chr> <dbl> <int> <dbl>
#> 1 1 tot_wss standa… 27830. 5 119.
#> 2 2 tot_wss standa… 18828. 5 815.
#> 3 3 tot_wss standa… 12454. 5 206.
#> 4 4 tot_wss standa… 10086. 5 207.
#> 5 5 tot_wss standa… 8865. 5 190.
#> 6 6 tot_wss standa… 8236. 5 404.
#> 7 7 tot_wss standa… 7211. 5 366.
#> 8 8 tot_wss standa… 6498. 5 365.
#> 9 9 tot_wss standa… 5920. 5 355.
#> 10 10 tot_wss standa… 5918. 5 338.
#> # … with 1 more variable: .config <chr>, and
#> # abbreviated variable names ¹num_clusters,
#> # ².estimator
#> # ℹ Use `colnames()` to see all variable namesThank You!
- Please use the package
- give us feedback
- make feature requests

github.com/EmilHvitfeldt/tidyclust