class: center, middle, title-slide # Logistic Regression ## AU STAT-427/627 ### Emil Hvitfeldt ### 2021-5-24 --- <div style = "position:fixed; visibility: hidden"> `$$\require{color}\definecolor{orange}{rgb}{1, 0.603921568627451, 0.301960784313725}$$` `$$\require{color}\definecolor{blue}{rgb}{0.301960784313725, 0.580392156862745, 1}$$` `$$\require{color}\definecolor{pink}{rgb}{0.976470588235294, 0.301960784313725, 1}$$` </div> <script type="text/x-mathjax-config"> MathJax.Hub.Config({ TeX: { Macros: { orange: ["{\\color{orange}{#1}}", 1], blue: ["{\\color{blue}{#1}}", 1], pink: ["{\\color{pink}{#1}}", 1] }, loader: {load: ['[tex]/color']}, tex: {packages: {'[+]': ['color']}} } }); </script> <style> .orange {color: #FF9A4D;} .blue {color: #4D94FF;} .pink {color: #F94DFF;} </style> # Classification Last we looked at regression tasks. In regression the response variable `\(Y\)` is quantitative In classification tasks, the response variable `\(Y\)` is **qualitative** This Difference will present some challenges we will cover this week --- .center[  ] --- # Examples of classification tasks - Should we sent an email ad to this person? - Are these symptoms indicative of cancer? - Given an image, which fruit is depicted? -- Two or more classes -- There can be uncertainty -- Can be more than one class at the same time --- # Classification visual <img src="index_files/figure-html/unnamed-chunk-2-1.png" width="700px" style="display: block; margin: auto;" /> --- # Classification visual - decision boundary <img src="index_files/figure-html/unnamed-chunk-3-1.png" width="700px" style="display: block; margin: auto;" /> --- # Classification visual <img src="index_files/figure-html/unnamed-chunk-4-1.png" width="700px" style="display: block; margin: auto;" /> --- # Classification visual - no hope <img src="index_files/figure-html/unnamed-chunk-5-1.png" width="700px" style="display: block; margin: auto;" /> --- # Nonlinear decision boundary <img src="index_files/figure-html/unnamed-chunk-6-1.png" width="700px" style="display: block; margin: auto;" /> --- # Logistic regression conceptually creates a linear line separating 2 classes Low flexibility, explainable method (we will talk about LDA, QLA, and K-nearest neighbors on Wednesday) --- # Logistic regression You might ask -- - Why can't you use linear regression? -- --- # Response encoding Propose we want to classify what kind of wine to market: - red - white `\(Y\)` has to be numeric for a linear model to work. -- We could decode `\(red = 0, white = 1\)`. -- but what would happen if we let `\(\hat{Y} >1\)` --- # Response encoding What if we have more than 2 classes? - red - white - .pink[rose] - .pink[dessert] - .pink[sparkling] We can't do `\(red = 1, white = 2, rose = 3, dessert = 4, sparking = 5\)` because there isn't natural ordering and nothing to indicate that dessert wine is twice of white wine --- # Logistic regression logistic (abstractly) models the probability that Y corresponds to a particular category -- Now some mathematics! --- # The Logistic Model We want to model the relationship between `\(p(X) = Pr(Y = 1|X)\)` and `\(X\)`. If we use a linear formulation `$$p(X) = \beta_0 + \beta_1X$$` then we will get negative probabilities which would be no good! --- # The Logistic Model .pull-left[ We need to restrict the values of `\(p(X)\)` to be between 0 and 1 We can use the .orange[logistic function] `$$f(x) = \dfrac{e^x}{1-e^x}$$` ] .pull-right[ <img src="index_files/figure-html/unnamed-chunk-7-1.png" width="700px" style="display: block; margin: auto;" /> ] --- # The Logistic Model Using the .orange[logistic function] gives us `$$p(X) = \dfrac{e^{\beta_0 + \beta_1X}}{1 + e^{\beta_0 + \beta_1X}}$$` Now no matter what the values of `\(X\)`, `\(\beta_0\)` or `\(\beta_1\)`, `\(p(X)\)` will always be contained between 0 and 1. --- # The Logistic Model If we start with `$$p(X) = \dfrac{e^{\blue{\beta_0 + \beta_1X}}}{1 + e^{\blue{\beta_0 + \beta_1X}}}$$` and we see that .blue[this] looks familiar, it is the linear combination we saw in linear regression we saw last week Explain what the parameter estimates mean --- # odds If we start with `$$p(X) = \dfrac{e^{\beta_0 + \beta_1X}}{1 + e^{\beta_0 + \beta_1X}}$$` after rearrangement gives `$$\dfrac{p(X)}{1 + p(X)} = e^{\beta_0 + \beta_1X}$$` --- # odds If we start with `$$p(X) = \dfrac{e^{\beta_0 + \beta_1X}}{1 + e^{\beta_0 + \beta_1X}}$$` after rearrangement gives `$$\orange{\dfrac{p(X)}{1 + p(X)}} = e^{\beta_0 + \beta_1X}$$` .orange[This] is called the **odds** and can take any value between 0 and `\(\infty\)`. --- # log-odds If we start with `$$p(X) = \dfrac{e^{\beta_0 + \beta_1X}}{1 + e^{\beta_0 + \beta_1X}}$$` after rearrangement gives `$$\dfrac{p(X)}{1 + p(X)} = e^{\beta_0 + \beta_1X}$$` taking the logarithm `$$\log\left(\dfrac{p(X)}{1 + p(X)}\right) = \beta_0 + \beta_1X$$` --- # log-odds `$$\blue{\log\left(\dfrac{p(X)}{1 + p(X)}\right)} = \beta_0 + \beta_1X$$` .blue[The left-hand side] is called the **log-odds** or **logit**. --- # How is this a classifier? Logistic regression is not modeling classes Logistic regression is modeling the probabilities that Y is equal on of the classes Logistic regression turns into a classifier by picking a cutoff (usually 50%) and classifying according to this threshold. --- # Logistic regression decision boundary <img src="index_files/figure-html/unnamed-chunk-8-1.png" width="700px" style="display: block; margin: auto;" /> --- # Non-linear separator <img src="index_files/figure-html/unnamed-chunk-9-1.png" width="700px" style="display: block; margin: auto;" /> --- # Coefficients Understanding: > Increasing `\(X\)` by one unit changes the log odds by a factor of `\(e^{\beta_1}\)` > The amount of change in `\(p(X)\)` depends on the current value of `\(X\)` --- # Making Predictions Fitting the model gives us `\(\hat{\beta_0}\)` and `\(\hat{\beta_1}\)` which we can use to construct `\(\hat{p}(X)\)` `$$\hat{p}(X) = \dfrac{e^{\hat{\beta_0} + \hat{\beta_1}X}}{1 + e^{\hat{\beta_0} + \hat{\beta_1}X}}$$` Plugging in the values of `\(\hat{\beta_0}\)`, `\(\hat{\beta_1}\)` and `\(X\)` gives us a prediction --- # Example with penguins <a href='https://allisonhorst.github.io/palmerpenguins'><img src='images/palmer.png' align="right" height="138.5" /></a> ```r library(palmerpenguins) penguins2 <- penguins %>% mutate(species = factor(species == "Adelie", labels = c("Adelie", "Not Adelie"))) library(parsnip) lr_spec <- logistic_reg() %>% set_engine("glm") %>% set_mode("classification") lr_fit <- lr_spec %>% fit(species ~ bill_length_mm + bill_depth_mm + body_mass_g, data = penguins2) ``` --- # Example with penguins <a href='https://allisonhorst.github.io/palmerpenguins'><img src='images/palmer.png' align="right" height="138.5" /></a> ```r lr_fit ``` ``` ## parsnip model object ## ## Fit time: 4ms ## ## Call: stats::glm(formula = species ~ bill_length_mm + bill_depth_mm + ## body_mass_g, family = stats::binomial, data = data) ## ## Coefficients: ## (Intercept) bill_length_mm bill_depth_mm body_mass_g ## 32.965109 -4.903438 8.616116 0.006746 ## ## Degrees of Freedom: 341 Total (i.e. Null); 338 Residual ## (2 observations deleted due to missingness) ## Null Deviance: 469.4 ## Residual Deviance: 9.652 AIC: 17.65 ``` --- # Example with penguins <a href='https://allisonhorst.github.io/palmerpenguins'><img src='images/palmer.png' align="right" height="138.5" /></a> ```r tidy(lr_fit) ``` ``` ## # A tibble: 4 x 5 ## term estimate std.error statistic p.value ## <chr> <dbl> <dbl> <dbl> <dbl> ## 1 (Intercept) 33.0 25.6 1.29 0.199 ## 2 bill_length_mm -4.90 2.65 -1.85 0.0647 ## 3 bill_depth_mm 8.62 4.81 1.79 0.0733 ## 4 body_mass_g 0.00675 0.00385 1.75 0.0800 ``` --- # Multi class classification We have so far only talked about what happens with 2 classes Logistic regression isn't able to work with multiple classes since it finds 1 best line to separate 2 classes --- # Logistic regression multiclass struggles <img src="index_files/figure-html/unnamed-chunk-13-1.png" width="700px" style="display: block; margin: auto;" /> --- # Logistic regression multiclass struggles <img src="index_files/figure-html/unnamed-chunk-14-1.png" width="700px" style="display: block; margin: auto;" /> --- # Evaluation To evaluate a classifier we need to quantify how good and bad it is performing 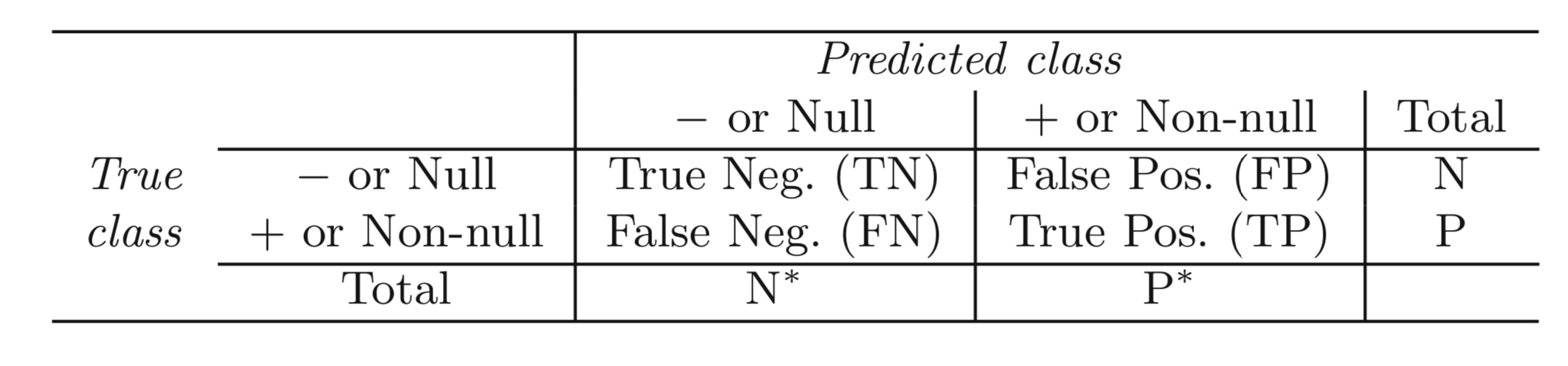 Different metrics will be different algebraic combinations of the above numbers --- # Evaluation metrics ## Accuracy `\(\dfrac{TN + TP}{TN + FN + FP + TP}\)` Percentage of correct predictions Drawback: If there are two classes A and B split 99% and 1%, you can get an accuracy of 99% by always predicting A --- # Evaluation metrics ## Sensitivity `\(\dfrac{TP}{FP + TP}\)` Defined as the proportion of positive results out of the number of samples that were positive --- # Evaluation metrics ## Specificity `\(\dfrac{TP}{FP + TP}\)` Measures the proportion of negatives that are correctly identified as negatives --- # ROC curve <img src="index_files/figure-html/unnamed-chunk-15-1.png" width="700px" style="display: block; margin: auto;" /> --- # Test-Train split We have spent some time talking about fitting model and measuring performance However, we need to be careful about how we go about that performance metrics calculated on the data that was used to fit the data is likely to mislead --- # Test-Train split In a prediction model, we are interested in the generalized performance. e.i. how well the model can perform on data it hasn't seen --- # Test-Train split <img src="index_files/figure-html/all-split-1.png" width="700px" style="display: block; margin: auto;" /> --- # Test-Train split We split the data into two groups (typically 75%/25%) - training data set - testing data set We do the modeling on the training data set (it can be multiple models) And then we use the testing data set **ONCE** to measure the performance --- # Why 75%/25%? There are no real guidelines as to how you split the data 80/20 split is also used It Will depend on data size --- # Why just once? If you are working in a prediction setting, the testing data set represents fresh new data If you modify your model you are essentially using information from the future to guide your modeling decisions This is a kind of data-leakage and it will lead to overconfidence in the model and will come back to bite you once you start using the model --- # How will I be able to iterate? We will talk more about how to efficiently use data in the next two weeks --- ## How should we handle unbalanced classes? <img src="index_files/figure-html/unnamed-chunk-16-1.png" width="700px" style="display: block; margin: auto;" /> --- ## How should we handle unbalanced classes? <img src="index_files/figure-html/unnamed-chunk-17-1.png" width="700px" style="display: block; margin: auto;" /> --- ## How should we handle unbalanced classes? <img src="index_files/figure-html/unnamed-chunk-18-1.png" width="700px" style="display: block; margin: auto;" /> --- # stratified sampling This stratification also works for regression tasks. The variable can be binned and samples to ensure equal distribution between training and testing data There is very little downside to using stratified sampling. --- # More Data Leakage Performing training-testing split in another place where data can leak Any transformation done to the data should be done **AFTER** the split occurs as to not have had future information affect the modeling process --- # rsample <a href='https://rsample.tidymodels.org/'><img src='images/rsample.png' align="right" height="138.5" /></a> **sample** provides functionally to perform all different kinds of data splitting with a minimal footprint --- # rsample example We bring back the `penguins` ```r penguins ``` ``` ## # A tibble: 344 x 8 ## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g ## <fct> <fct> <dbl> <dbl> <int> <int> ## 1 Adelie Torgersen 39.1 18.7 181 3750 ## 2 Adelie Torgersen 39.5 17.4 186 3800 ## 3 Adelie Torgersen 40.3 18 195 3250 ## 4 Adelie Torgersen NA NA NA NA ## 5 Adelie Torgersen 36.7 19.3 193 3450 ## 6 Adelie Torgersen 39.3 20.6 190 3650 ## 7 Adelie Torgersen 38.9 17.8 181 3625 ## 8 Adelie Torgersen 39.2 19.6 195 4675 ## 9 Adelie Torgersen 34.1 18.1 193 3475 ## 10 Adelie Torgersen 42 20.2 190 4250 ## # … with 334 more rows, and 2 more variables: sex <fct>, year <int> ``` --- # rsample example Use `initial_split()` from rsample to generate a `rsplit` object ```r set.seed(1234) # remember the seed! penguins_split <- initial_split(penguins) penguins_split ``` ``` ## <Analysis/Assess/Total> ## <258/86/344> ``` This object store the information of what observations belong to each data set --- # rsample example `training()` and `testing()` is used to extract the training data set and testing data set ```r set.seed(1234) # remember the seed! penguins_split <- initial_split(penguins) penquins_train <- training(penguins_split) penquins_test <- testing(penguins_split) dim(penquins_train) ``` ``` ## [1] 258 8 ``` ```r dim(penquins_test) ``` ``` ## [1] 86 8 ```